Understanding Marker Genes in Bacterial Genomes
Written on
Chapter 1: Marker Genes and Their Importance
Marker genes serve as vital indicators in genomics, particularly within bacterial genomes. Just as a marker pinpoints a location on a map, these genes help identify and classify organisms in metagenomic studies. This article delves into the concept of marker genes, their applications, and offers insight into a widely used gene prediction tool.

What Exactly Are Marker Genes?
According to Wikipedia, a genetic marker is defined as a gene or DNA segment with a known chromosomal location that aids in the identification of individuals or species. Due to mutations and variations in the genome, these markers can differ based on their composition and position.
Single-Copy Marker Genes
In bacterial cells, single-copy marker genes are typically found only once per cell. This means that each bacterial cell contains a solitary copy of these essential genes, which are crucial for their survival and can be identified across a wide range of bacterial species.
Research has previously focused on pinpointing marker genes capable of distinguishing closely related organisms. Notably, protein-coding marker genes that are infrequently transferred horizontally and exist in single copies within genomes have been identified, including sets of 40 and 107 marker genes.
Usage of Marker Genes
Marker genes play a significant role in taxonomic profiling of environmental samples, assisting in the identification of gene families. They are also utilized in phylogenetic studies to trace the evolutionary lineage of organisms.
Recent advancements have led to reference-free binning tools, like MaxBin and SolidBin, which leverage single-copy marker genes to ascertain species richness in various samples. Additionally, tools such as MyCC utilize these genes to refine the resulting clusters.
Gene Prediction Tools
To extract marker genes, various gene prediction tools are employed. Some notable examples include:
- Glimmer
- MetaGene
- GeneMark
- FragGeneScan
- fetchMG
Example of Using FragGeneScan
Let’s explore how to use FragGeneScan for gene prediction. You can download FragGeneScan from:
Follow the README instructions to compile and execute FragGeneScan. Below are the parameters and options you will encounter.
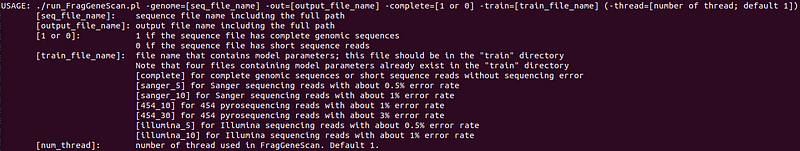
Once you have your complete genomic sequence, you can run FragGeneScan as follows:
./run_FragGeneScan.pl -genome=<sequence_file> -out=<output_file> -complete=1 -train=complete -thread=<num>
If you are working with assembled contigs, the command would be:
./run_FragGeneScan.pl -genome=<contigs_file> -out=<output_file> -complete=0 -train=complete -thread=<num>
FragGeneScan will produce four output files:
- <output_file>.out: Coordinates of predicted genes
- <output_file>.fnn: Nucleotide sequences of the predicted genes
- <output_file>.faa: Amino acid sequences of the predicted genes
- <output_file>.gff: Gene prediction results
With the <output_file>.faa, you can utilize HMMER to identify single-copy marker genes within the sequences.
Final Thoughts
Marker genes have emerged as a powerful tool in bioinformatics, enabling researchers to gain valuable insights into the taxonomic classification and evolutionary history of bacterial and archaeal species. The field of metagenomics greatly benefits from research focused on these genes.
I hope you find this article informative and encourage you to experiment with the tools discussed.
Cheers, and stay safe!
References
[2] Ciccarelli et al. (2006) Toward Automatic Reconstruction of a Highly Resolved Tree of Life, SCIENCE 03: 1283–1287
Chapter 2: Video Insights into Marker Genes
In this informative video titled "Marker Gene Based Analysis," viewers can learn about the fundamentals and applications of marker genes in genomic studies.
The second video, "Analysis of Metagenomic Data: Introduction to Metagenomics," provides a deeper understanding of metagenomics and the role of marker genes in this field.